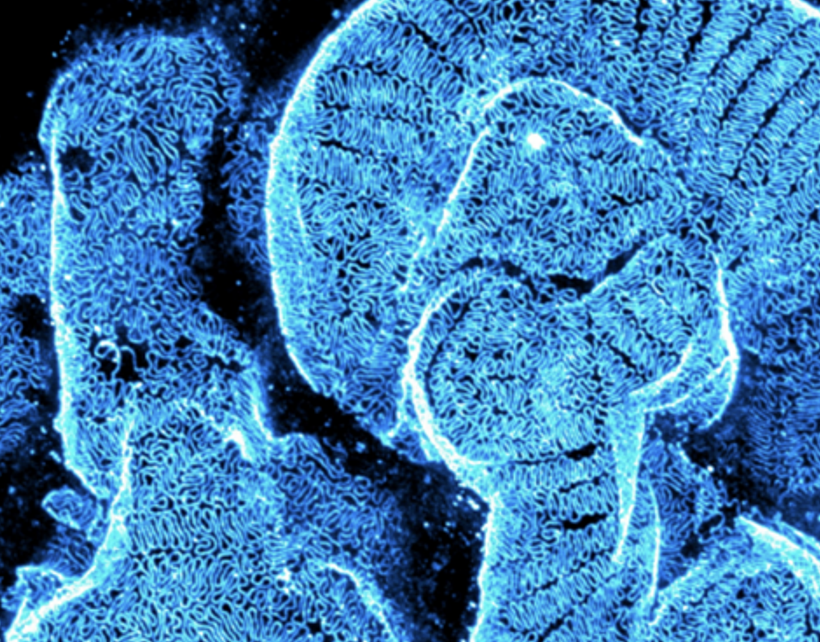
About the project
Objective
Kidney diseases affects approximately 10% of the world’s population and is projected to become one of highest cause of life-lost-years within two decades. Despite this growing healthcare threat, diagnostic methods remain insufficiently precise, which unfortunately hinders early quantitative kidney disease staging. We aim to demonstrate a precision medicine pipeline that combines high-resolution optical 3D whole biopsy imaging with advanced AI-assisted 3D image analysis. Our approach eliminates preparation-induced biases by not slicing tissue biopsies, thereby preserving native 3D architecture and in-context spatial relationships. High-resolution imaging in 3D will spatially resolve key structural and molecular disease markers across scales in optically cleared biopsies. Moreover, our pipeline will automate 3D disease staging using AI-based image analysis, employing cutting-edge self-supervised learning to extract, segment, and quantify disease-relevant features objectively.
Background
Pathology impacts all aspects of patient’s care from identifying a disease, to monitoring its progression and to make crucial treatment decisions. Clinical kidney pathology thus plays a central role as method for diagnostic support in healthcare to help renal physicians and patients. Present nanoscale kidney pathology analysis is done in ultra-thin biopsies by electron microscopy, losing precious in-context 3D information of morphological and molecular pathophysiology. Moreover, used workflows are highly labor intense across sample preparation, imaging and imaging analyzing steps.
Cross-disciplinary collaboration
Imaging and image analysis are inherent pathology tools that can deliver diagnostic support and guide patient care. Our developed kidney pathology workflow will be faster, more precise, save healthcare labor costs, and be less biased with automate AI analysis support. The exploration of self-supervised methods for functional structure analysis is furthermore a rapidly growing and impactful area in AI, as researchers seek more sophisticated ways to analyze high-dimensional biological data without manual annotations. Our work positions KTH at the forefront of this cutting-edge field, meeting the rising demand for robust, scalable AI tools that can improve diagnostic accuracy and consistency.
PI: Hans Blom, Applied Physics/SCI/KTH
Co-PI: Gisele Miranda, Computer Science/EECS/KTH